Serological relationships among strains of grapevine leafroll-associated virus 4 reflect the evolutive behavior of its coat protein gene
DOI:
https://doi.org/10.48162/rev.39.100Keywords:
molecular characterization and serology, grapevine leafroll disease, ampelovirus, selection pressure, antigenic propertiesAbstract
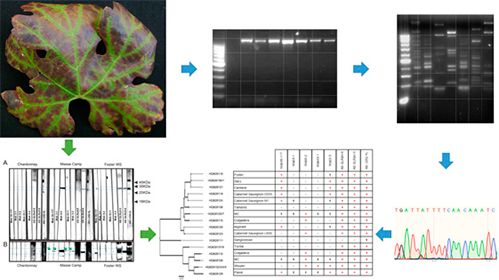
This research studied serological relationships and genetic diversity of Argentinean isolates of grapevine leafroll-associated virus-4 (GLRaV-4). Phylogenetic analysis of coat protein (CP) sequences from 19 local isolates revealed clustering with the previously described GLRaV-4 strain 5, strain 6, and strain 9 groups. Evolutionary sequence analysis of the obtained and database-available sequences showed evidence of recombination events. Additionally, both CP N- and C-terminal regions appeared to be under purifying selection, but the N-terminal region presented seven sites under positive selection, with a dN/dS ratio 5-fold greater than that of the C-terminal region. Serological reactivity against monoclonal antibodies supports a higher occurrence probability for linear epitopes in the N-terminal region, as inferred by the sequence analysis. The obtained results reflect an unusual evolutionary behavior of the CP that, together with protein serological reactivity, suggests biological significance of the observed variability.
Highlights:
- The occurrence of GLRaV-4 strains -5, -6 and -9 was recorded in Argentina.
- The coat protein sequence showed a highly variable N-terminal region among the strains.
- An evolutionary analysis inferred the occurrence of sites under positive diversifying selection pressure.
- The most variable region of the coat protein presented the highest probability of occurrence of linear epitopes.
Downloads
Published
Issue
Section
License
Copyright (c) 2018 Revista de la Facultad de Ciencias Agrarias UNCuyo

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License.
Aquellos autores/as que tengan publicaciones con esta revista, aceptan las Políticas Editoriales.


