Development and characterization of SSR markers for Trichloris crinita using sequence data from related grass species
Keywords:
Chloridoideae, orage grass, genetic diversity, marker transferability, microsatellitesAbstract
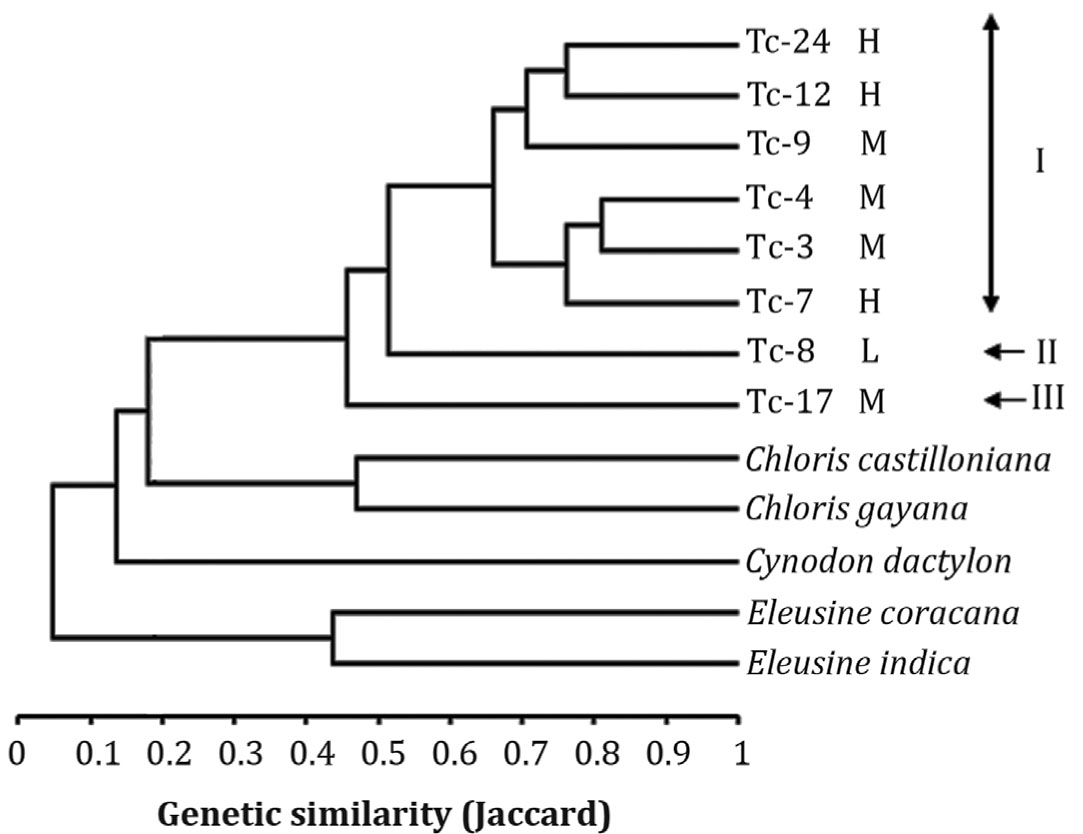
Trichloris crinita is among the most important native forage grasses in arid regions of America. Despite its importance, molecular resources and sequence data are extremely scarce in this species. In the present study, SSR markers were developed using available DNA sequences from grass taxa phylogenetically-related to Trichloris (Eleusine coracana, Cynodon dactylon and ‘Cynodon dactylon x Cynodon transvaalensis’). Marker transferability was evaluated in a panel of eight T. crinita accessions and five closely-related species. Of the 105 SSR primer pairs evaluated, 16 amplified products of expected size in T. crinita, whereas transferability to other grass species ranged from 12 (in Chloris castilloniana) to 28 SSRs (in Eleusine coracana). Six of the 16 SSR markers successfully transferred to T. crinita (37.5%) were polymorphic, and were further used to assess genetic diversity in eight T. crinita accessions. The analysis revealed a total of 23 SSR alleles (3.83 alleles/locus), allowing the discrimination of all T. crinita accessions, with pair-wise genetic similarities ranging from 0.35 to 0.81 (Jaccard coefficient). Mean (and range) values for observed (Ho) and expected heterozygosity (He) were 0.53 (0.0-1.0) and 0.63 (0.48-0.79), respectively.
Downloads
Published
Issue
Section
License
Aquellos autores/as que tengan publicaciones con esta revista, aceptan las Políticas Editoriales.


